methyl-DNAshape: High-throughput prediction server for methylated DNA shape features
This manual will walk you through the steps involved in using methyl-DNAshape web server. Documentation in Read the Docs version is available here.
Step 1:
- Please visit methyl-DNAshape and click on the
 link on the main menu, or you can directly visit the prediction page here.
link on the main menu, or you can directly visit the prediction page here.
Step 2:
Input:
- Single or multiple (FASTA format) sequences (in the left box). The input sequence is case insensitive.
- Indices of CpG in sequence(s) needs to be converted to Mpg (in the right box)
- Upload a file containing sequence(s) in FASTA format (for example, see sample.fasta)
- Upload corresponding position indices file (for example, see pos.fasta)
OR
-
Output format (optional):
- delimiter (feature values to be separated by)
- Entries per line (#values per line)
NOTE: Where the symbol M or the position number always refers to cytosine of methylated CpG on the leading strand (5'-3'),
the cytosine on lagging strand will also be considered methylated. CpG and Mpg refers to CG and Mg respectively in sequences.
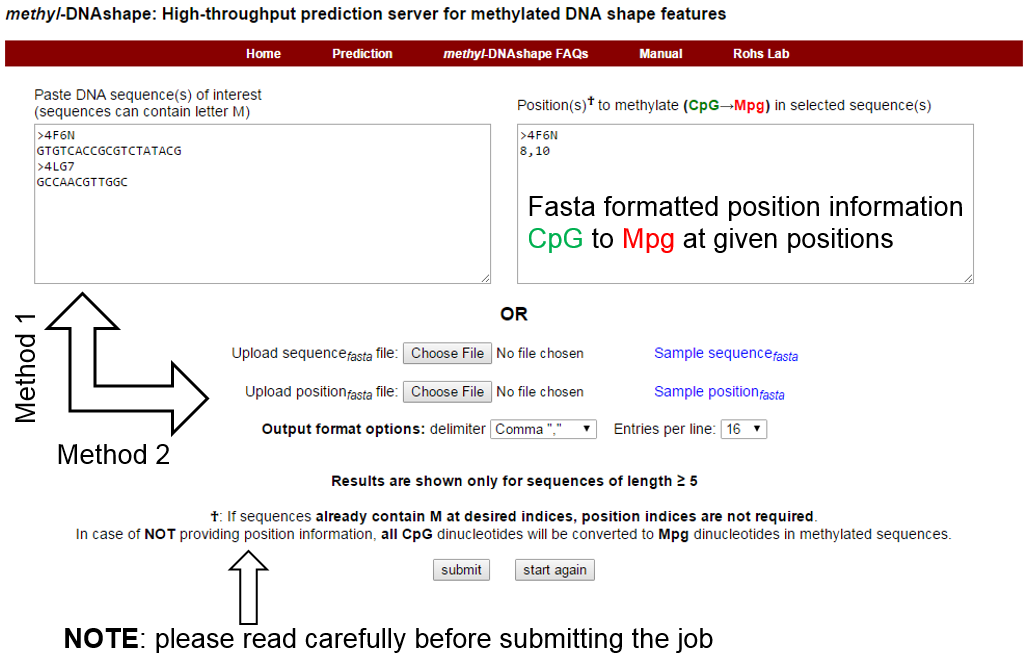
Position textbox in the figure above contains indices only for sequence 4F6N; this implies that sequence 4LG7 will be treated as is. The figure below illustrate the same:
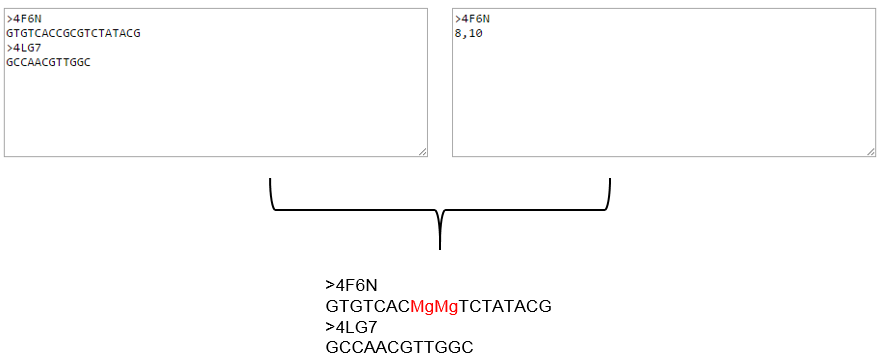
We can see that CpG dinucleotides at mentioned indices (8, and 10) are converted to Mpg dinucleotides (highlighted in red) in 4F6N and there are no changes in 4LG7.
Step 3:
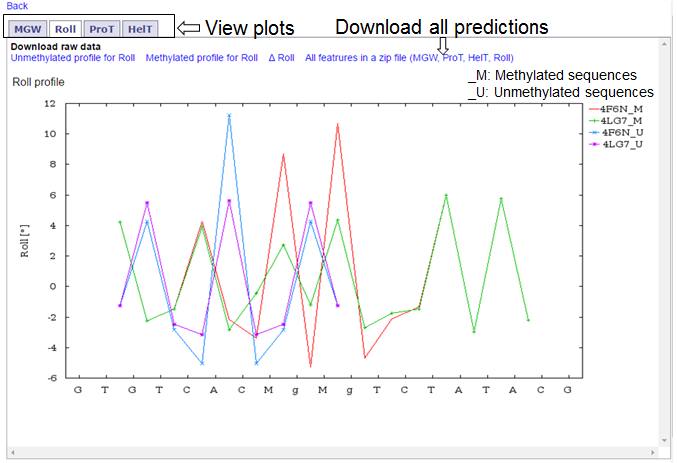
- Once the program has run, you will be able to see the plots and download links for results. You can view different plots by navigating through tabs titled MGW, ProT, Roll and HelT. All results can be downloaded in a zip file. Please see the image below.