methyl-DNAshape: High-throughput prediction server for methylated DNA shape features
Q1. What is methyl-DNAshape?
Answer: methyl-DNAshape is a web server for the high-throughput prediction of methylated DNA structural features. It predicts DNA shape features such as minor groove width (MGW), propeller twist (ProT), Roll, and helix twist (HelT) for a given DNA sequence.
Q2. Why methyl-DNAshape is needed?
Answer: In general proteins employ two types of readout mechanisms while binding to DNA, base and shape. In the earlier, the chemical signature of sequences are important and in the latter, the structural. We previously developed DNAshape, a high-throughput method for predicting shape profile of DNA which helps better understand the shape readout. But, epigenetic modification on DNA, for example CpG methylation, was due. methyl-DNAshape addresses the same.
Q3. What are these structural features and why are they important?
Answer:The following figure is schematic explanation of these structural features. They play important roles in DNA shape readout.
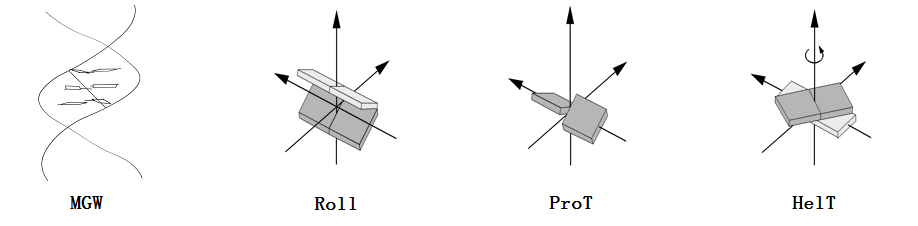
Q4. What is the input format for the methyl-DNAshape web server?
Answer: Please see the manual here. By default, the web server changes all CpG dinucleotide(s) to Mpg where M represents a methylated cytosine. Please replace CpG by Mpg manually in order to get position-specific results in cases where not all CpG steps are methylated.
Q5. What is the sequence length limit?
Answer: The sequence should be at least 5 base pairs in length. There is no limit on the maximum sequence length, but if the sequence length is greater than 1 Mb (1 million base pairs), then please contact us.
Q6. What are Δ feature (MGW, ProT, HelT and Roll) values?
Answer: The Δ feature values are calculated by subtracting the unmethylated feature value from methylated feature value for each nucleotide position of a given sequence.
Q7. What are letters "M" and "g" in sequences representing?
Answer: The letter "M" represents methylated cytosine and the letter "g" represents guanine next to "M". We needed letter "g" to distiungish regular guanine letter letter "G" to address the methylation context.
Q8. Why do I see only one sequence as ticks on the X-axis?
Answer: In order to not make the plots very crowded, we only show one sequence (the longest of input sequences) on the X-axis as ticks in an unit interval.